Identification of selective inhibitors of the NLRP3 inflammasome
Abstract
The innate immune system is the host’s first line of defense, it acts thanks to the presence of pattern-recognition receptors (PRR) that recognize the presence of pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs). Among these receptors, Nod-like-receptors (NLRs) play an important role. NLRP3 is the most studied among the NLRs.
Upon its activation, the NLRP3 protein assembles with the ASC adapter and pro-caspase-1 forming the NLRP3 inflammasome complex. The active inflammasome induces the activation of caspase-1 with consequent maturation of the pro-inflammatory cytokines IL-1β and IL-18. Caspase-1 also induces the cleavage of gasdermin D, which translocates into cell membranes, where it induces the formation of pores promoting unconventional protein secretion and cell death by pyroptosis.
Aberrant activation of NLRP3 is found in Cryopyrin-Associated Autoinflammatory Syndromes (CAPS), it has been associated with sterile inflammation and contributes to several chronic diseases, including neurodegenerative diseases, atherosclerosis and type II diabetes.
At the moment, no selective NLRP3 inhibitors have made to the clinique.
The project aims at discovering novel selective inhibitors of NLRP3 through a joint effort of the following research groups:
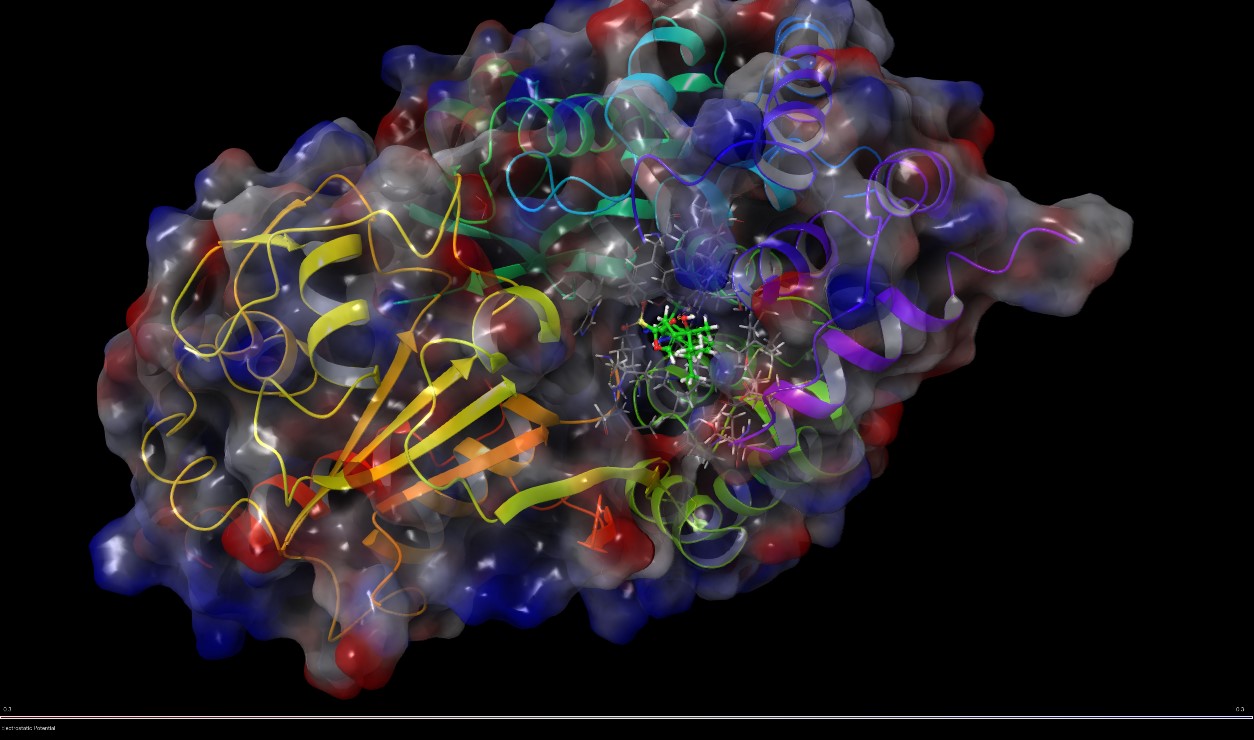
Molecular Informatics: The group is charge for virtual screening campaigns and binding mode rationalization for small molecules. It also assists Med Chem group providing models to be used for molecular optimization.
Medicinal Chemistry: The medicinal chemistry platform supports the early stages of screening campaigns with structure validation and hit confirmation studies and with structure-activity relationship studies in the later hit-to-lead phase.
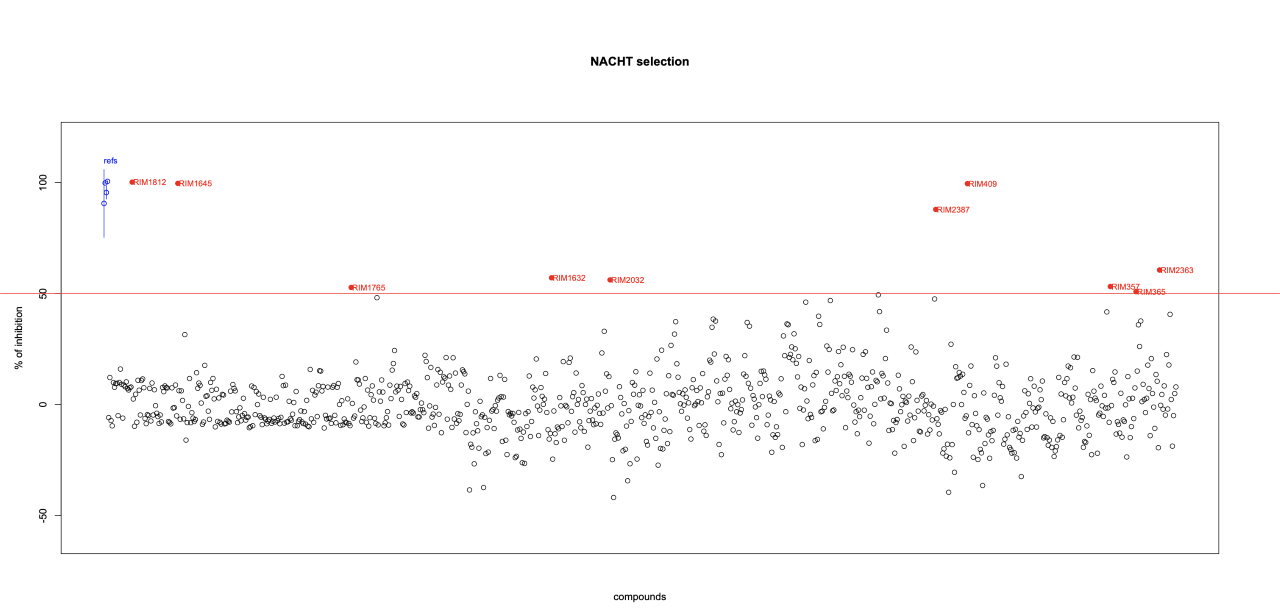
Advanced Data Analysis: Within this project, the Advanced Data Analysis group covers the biostatistical and data management issues. Specifically, it is on charge of the design of the plates, i.e., according to the existing constrains (number of compounds to be tested, number of reference compounds, plate type and number of experimental replicates needed), it assigns how the HTS plates has to be populated . Then, after the plates have been processed through the experimental protocols, we collect, preprocess and analyze all the data and provide statistical significance of the experimental activity of each screened compound. We are also developing automated workflows devoted to the most frequent data analysis activities involved in the HTS lab, in order to automate the data analysis processes.
Structural biology and biophysics:
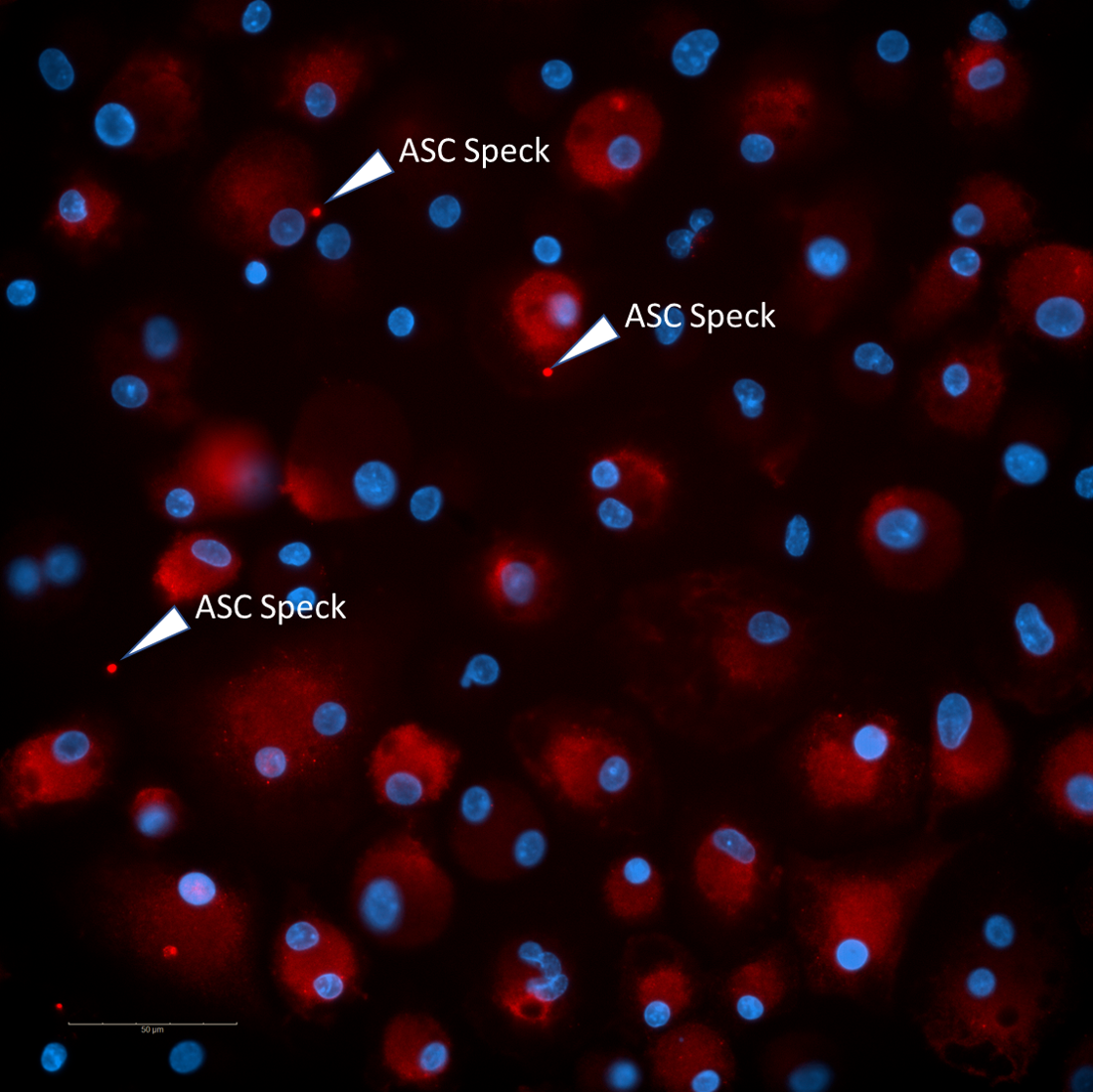
Screening Lab: We test libraries of compounds using a primary phenotypic assay where the release of IL-1b after canonical NLRP3 inflammasome activation is evaluated in a human monocyte-macrophage cell line (THP1). Primary actives are then evaluated through a number of orthogonal and secondary assays that evaluate pyroptosis (LDH release), caspase-1 activity (enzymatic assay), ASC speck formation, membrane pore formation, off-target effects and toxicity.
Pipeline
-
CLINICAL
NEED -
DISEASES
ANALYSIS - DISCOVERY
-
PRECLINICAL
VALIDATION -
PRECLINICAL
DEVELOPMENT -
CLINICAL
STUDIES
Principal Investigator
Contatto
Aree terapeutiche:
Prodotto:
Biomarcatori – Dispositivi biomedicali e Organi artificiali
Collaborazioni:
Institut de la Vision, Sorbonne Universitè (France)
Institute of Innate Immunity, University Hospital Bonn (Germany)
Scarica il pdf del progetto