Piattaforma di Proteomica
Description
Ri.MED hosts a state-of-the-art proteomics platform designed to support high-resolution quantitative proteomics and translational research applications. The platform comprises the following instrumentation.
Instrumentation for LC–MS/MS analysis
-
Astral mass spectrometer (Thermo Fisher Scientific) coupled to a Vanquish Neo uHPLC system
-
Exploris 480 mass spectrometer (Thermo Fisher Scientific) coupled to a Vanquish Neo uHPLC system
-
Q Exactive mass spectrometer (Thermo Fisher Scientific) coupled to a high-performance nanoLC system (UltiMate 3000 RS)
Instrumentation for sample preparation
-
BeatBox tissue homogenizer (PreOmics)
-
Dedicated instrumentation for major proteomic sample-processing workflows, including in-gel digestion, SP3, and FASP methodologies
Hardware and software for data analysis
-
Two dedicated workstations and NAS systems for big-data analysis and long-term storage
-
Proteomics data-analysis software, including Spectronaut, Proteome Discoverer, DIA-NN, Perseus, and additional specialized bioinformatics tools
Instrumentation for orthogonal validation of proteomics data
-
Biosafety cabinets and instrumentation for cell and tissue culture
-
Platforms for ELISA assays and Western blotting
Platform activities
The instrumentation available within the platform enables chromatographic separation of peptides generated by proteolytic digestion of complex protein systems, such as cell lysates, conditioned media, tissues, or biological fluids. Peptides are subsequently ionized by electrospray and fragmented into ions with distinct mass-to-charge ratios, generating mass spectra that are unique to each peptide.
Mass spectra are analyzed using dedicated proteomics software, allowing the identification of individual proteins present in the original biological sample. In addition, the platform supports advanced quantitative proteomics analyses, making it possible not only to identify proteins but also to accurately quantify protein expression levels across different biological or experimental conditions.
The overarching goal of the Ri.MED proteomics platform is to provide high-quality quantitative proteomics analyses in support of scientific research, both within the Institute and for national and international research groups. Through these activities, Ri.MED aims to establish itself as a reference center for advanced proteomics research at the regional, national, and international levels.
Expertise
- Concentrazione di proteine da terreni condizionati;
- Misura spettrofotometrica (Bradford, BCA, micro BCA);
- Precipitazione e analisi chimica del campione;
- Digestione triptica in gel ed in soluzione;
- Preparazione di peptidi con metodo FASP;
- STAGE (STop And Go Extraction) Tip;
- Sample CleanUp;
- Frazionamento in base al pH;
- Secretome protein enrichment with click sugars (SPECS);
- Proteomica quantitativa LFQ;
- Western Blot;
- SDS-PAGE;
- Analisi qualitativa e quantitativa di proteine mediante cromatografia liquida/spettrometria di massa per proteomica “Bottom Up” e “Shot-gun”.
Facilities & Equipment
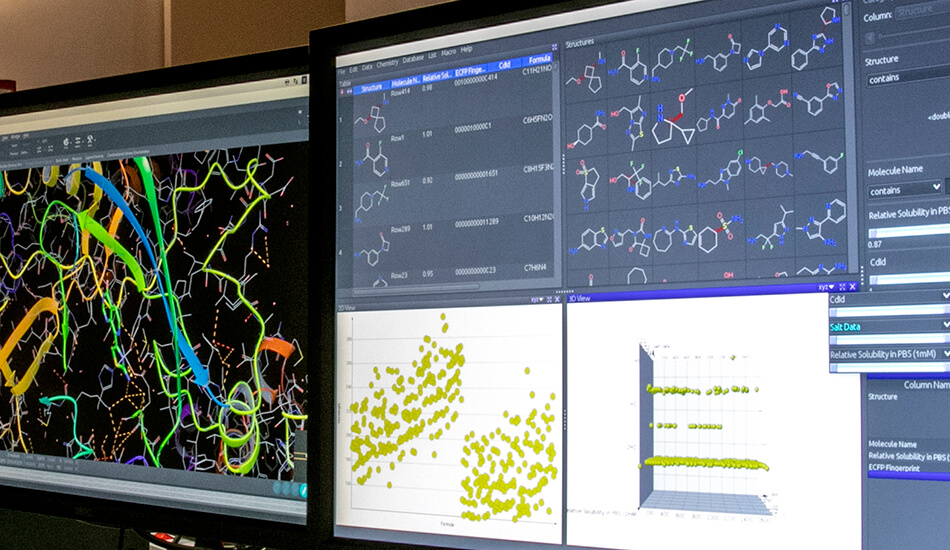
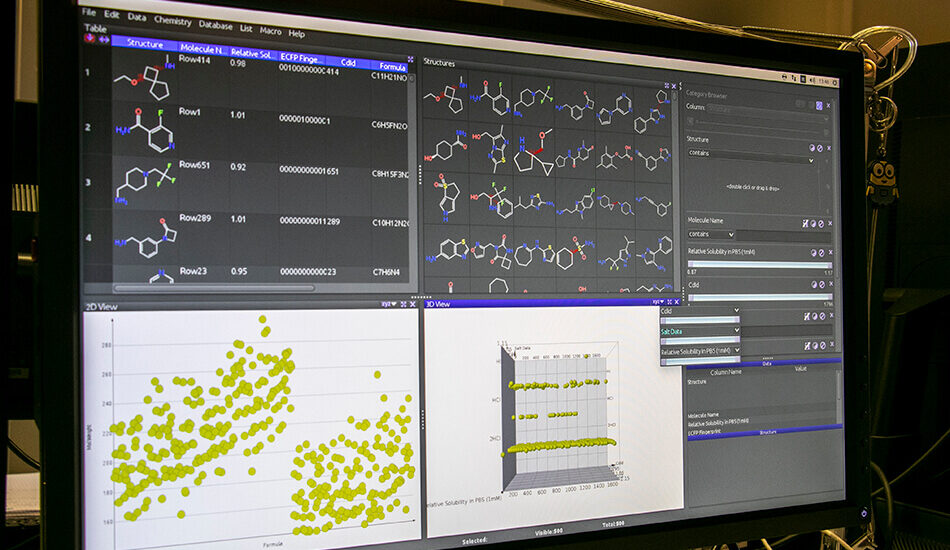
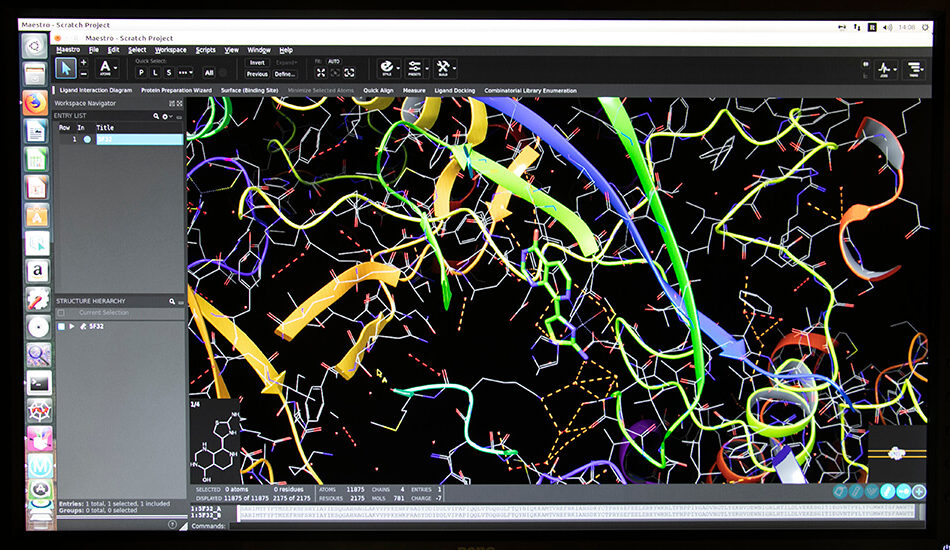
Software
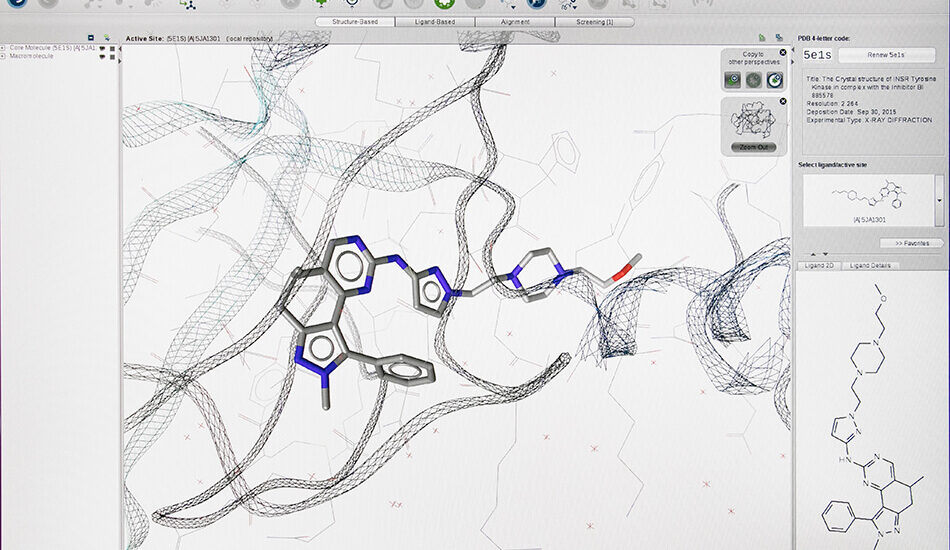
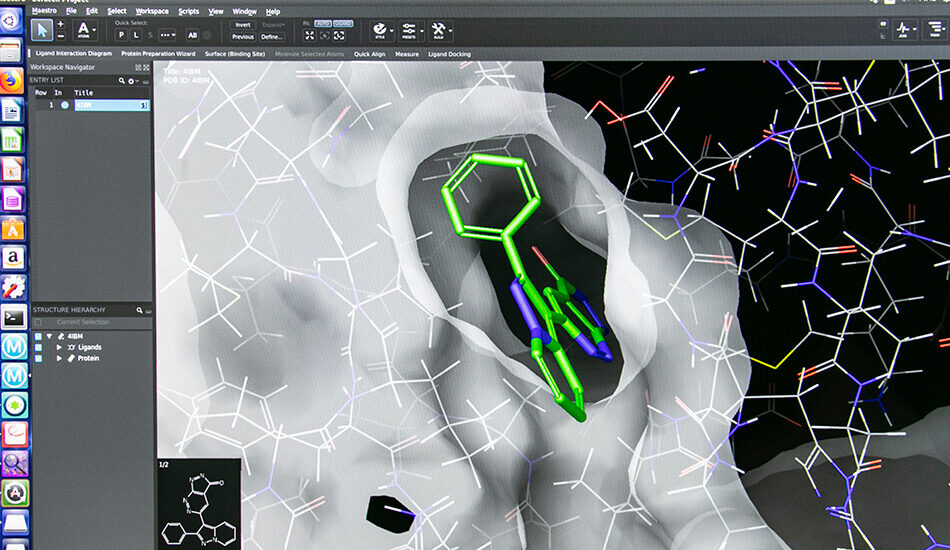
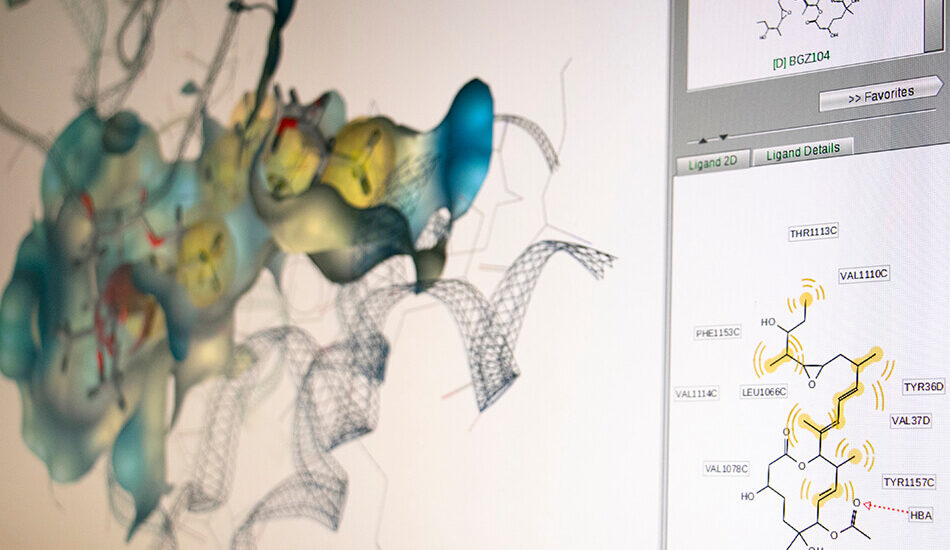
- Schrodinger suite for small molecule drug discovery;
- LigandScout expert suite;
- Autodock and Autodock Vina;
- Desmond (OPLS2005 and OPLS3e);
- AMBER;
- NAMD;
- VMD;
- Gromacs;
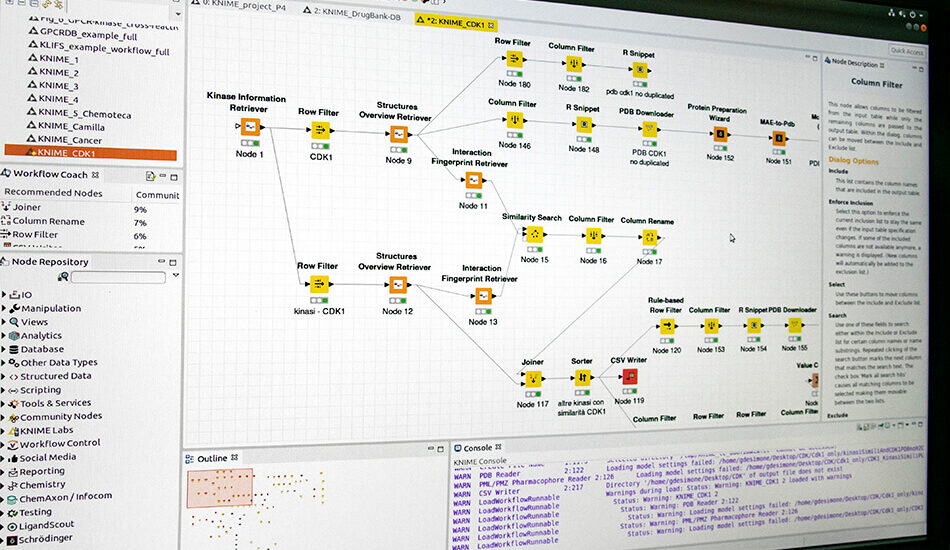
- RDKit;
- KNIME.
Hardware
- 6 Workstations;
- Server in HPC mode: 200 CPUs e 2 x NVIDIA Tesla K80.
Capacità di calcolo:
- Library optimisation → ~ 6,000 molecole/min
- Virtual Screening HTVS → ~ 5,000 molecole /min
- Virtual Screening SP → ~ 1,500 molecole /min
- Molecular Dynamics → ~ 200 ns/giorno/scheda (su un sistema medio di 40,000 atomi)
Piattaforma integrata in silico
Il gruppo sta realizzando una piattaforma integrata per lo studio dei network molecolari in collaborazione con il gruppo di bioinformatica.
Contacts:
Collaboration:
- Institute for Aging and Chronic diseases, University of Liverpool, Regno Unito
- German Center for Neurodegenerative Diseases (DZNE), Monaco, Germania
- Queen Mary University of London, Regno Unito
- Dipartimento di Scienze Farmacologiche, Università di Pisa, Italia
- STEBICEF, Università di Palermo, Italia
Hardware
I am text block. Click edit button to change this text. Lorem ipsum dolor sit amet, consectetur adipiscing elit. Ut elit tellus, luctus nec ullamcorper mattis, pulvinar dapibus leo.