Development of new tools for the fight against SARS-CoV-2
Brief description
The COVID-19 pandemic has led to the development of various strategies to prevent infection or alleviate clinical manifestations. Until now, most of the efforts have been invested in the development of vaccines, capable of inducing an immune response against the viral protein Spike, and neutralizing antibodies against Spike. However, considering the mutant nature of SARS-CoV-2 which mainly affects the Spike region, the effectiveness of the tools available to date is not demonstrated for current and future variants of the virus.
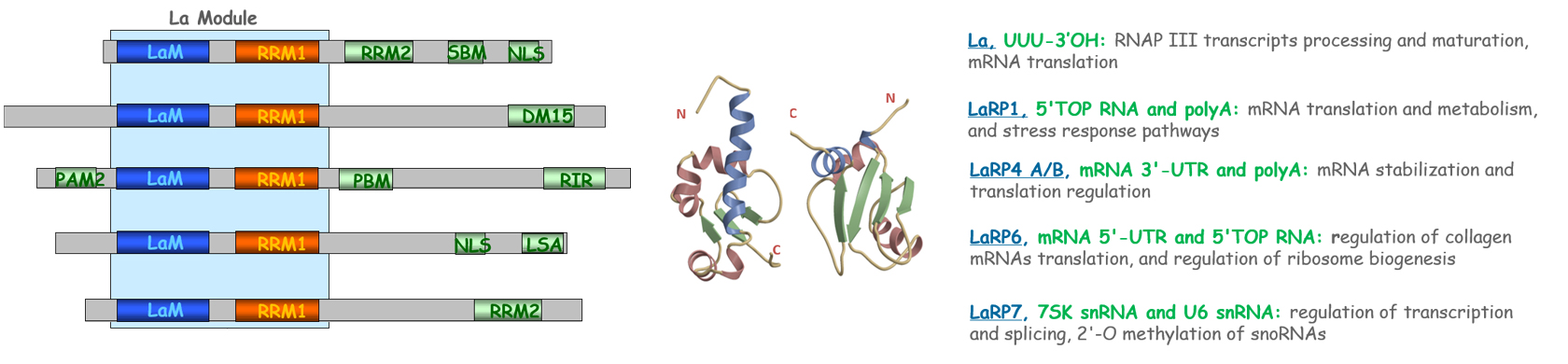
Moreover, the infection of SARS-CoV-2 causes a storm into the homeostasis of host cells due to the hijacking of a plethora of host factors required to complete the virus lifecycle. Several host proteins have been found to be involved in Protein-Protein interactions with the viral Replication Transcription Complex (RTC) composed by three viral non-structural proteins (nsps): nsp12 (RNA-dependent-RNA-polymerase), that mediates the nucleotide polymerization, and nsp8 and nsp7, acting as primase complex. In particular, findings based on proteomic studies and computational models suggested that RNA-binding proteins such as La-Related Proteins (LaRPs), a superfamily of proteins characterized by an extremely conserved N-terminal motif (La Module), related to the cellular RNAs regulation and involved in the viral genome replication.
Our studies are focused on the one hand to the biophysical and structural characterization of possible interactions that involve the pivot triad nsp12-8-7 of the RTC with host factors LaRPs, to determine if one or more members of this peculiar superfamily is crucial for viral replication. By providing kinetic and structural information, by using structural and biophysical techniques, such as Bio-Layer Interferometry (BLI), Isothermal Titration Calorimetry (ITC), Nuclear Magnetic Resonance (NMR), and cryo-Electron Microscopy (cryo-EM), our research aims to contribute for developing new therapeutic approaches.
On the other hand, the goal of our research project is to find more durable and broad-spectrum treatments that are effective against the numerous variants of the virus and that can also counteract future coronavirus pandemics. In particular, we are involved in the development of antibodies, in the form of single chain fragment variable (scFv), against the Nucleocapsid protein (N) and the Nsp9 protein of SARS-CoV-2, rather than on the Spike protein whose rapid and high mutability results in a reduction in the effectiveness of the treatments available. Both chosen proteins are of extreme biological and therapeutic relevance. Nsp9 is a highly conserved ssRNA-binding dimeric protein among Betacoronaviruses and is involved in virus replication and transcription. Protein N, also dimeric, packages the positive strand of viral RNA and plays an important role in improving the transcription efficiency of subgenomic viral RNA.
Impact
Since the onset of the COVID-19 pandemic, most efforts have been invested in developing vaccines capable of inducing an immune response against the viral protein Spike, and neutralizing antibodies against Spike. However, one of the questions that researchers immediately tried to answer concerns the extreme variability of the SARS-CoV-2 virus, mainly due to the highly mutability nature of the Spike protein, which makes the therapeutic tools developed so far less and less effective. Therefore, it remains essential to investigate other stages of viral infection and replication, as well as the molecular mechanism by which viruses abduct and exploit host factors. In this context, a deepen knowledge of the role that host proteins play during the viral replication will contribute to figure out the molecular bases behind the infection of coronaviruses.
We also aim to develop new molecules against proteins other than Spike, while implementing a platform and workflow for rapid and decisive intervention in the event of future coronavirus pandemics. The data produced so far on Nsp9 and the Nucleocapsid protein N provide the prerequisite for selecting and developing new neutralizing antibodies against these two proteins, thus preventing viral replication and/or attenuating the aggressiveness of the virus.
Pipeline
-
CLINICAL
NEED -
DISEASES
ANALYSIS - DISCOVERY
-
PRECLINICAL
VALIDATION -
PRECLINICAL
DEVELOPMENT -
CLINICAL
STUDIES
Principal Investigator
Contact
Team di progetto:
Elisa Monaca
Therapeutic Areas:
Product:
Drugs – Biologics
Collaborations:
- Randall Division of Cell and Molecular Biophysics, King’s College London (KCL), UK
- EBRI – European Brain Research Institute Rita Levi-Montalcini, Rome, Italy
- International Covid-19 NMR Consortium
Scarica il pdf del progetto
