Development of Automatic Fully 3D Algorithms for Biomedical Image Segmentation and Classification
Abstract
In the field of biomedical imaging, target segmentation is routinely used as the first step in any automatized disease diagnosis system (i.e. radiotherapy system) and, in the last few years, in radiomics studies to obtain great volumes of quantitative data from medical images. These data are then used as imaging biomarkers to identify any possible associations with patient outcome. The first step of a radiomics workflow is the target (e.g. tumor or organ) delineation in such a way as to avoid distortions in parameter extraction. Although manual delineation seems like the most intuitive and easily implemented way of obtaining target volume, it is a time consuming process and it is subject to the greatest inter- and intra-observer variability. This variability causes irreproducible results in the radiomics signature that is highly influenced by the region of interest drawn to identify the tumour. For this reason, an automatic and operator-independent target delineation method is mandatory.
Impact:
The segmentation process remains a popular and challenging area of research. Nowadays clinical activity places a high level of demand on segmentation algorithms, which are required to produce repeatable results, to be independent by the choices performed by the user and capable of processing in real-time. The project proposes a segmentation system specifically engineered to reach the maximum level of automation and capable of obtaining an operator independent segmentation. In the specific, we propose two methodology:
– A fully 3D active surface (AS) driven by a 3D machine learning component (i.e., 3D tissue classification) for segmentation of the tumours in brain districts in PET;
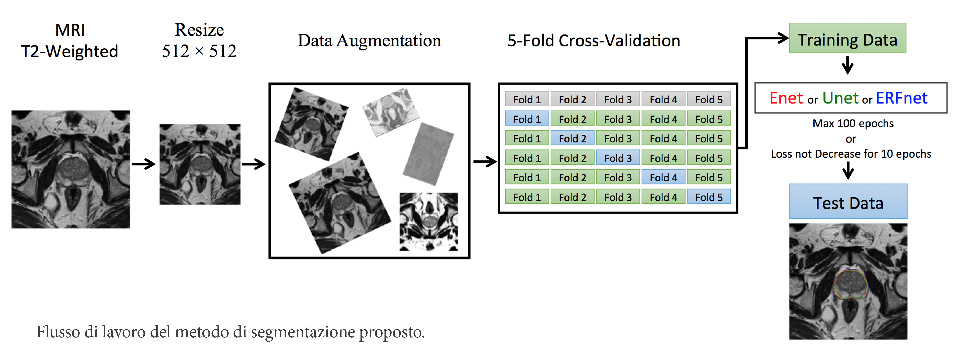
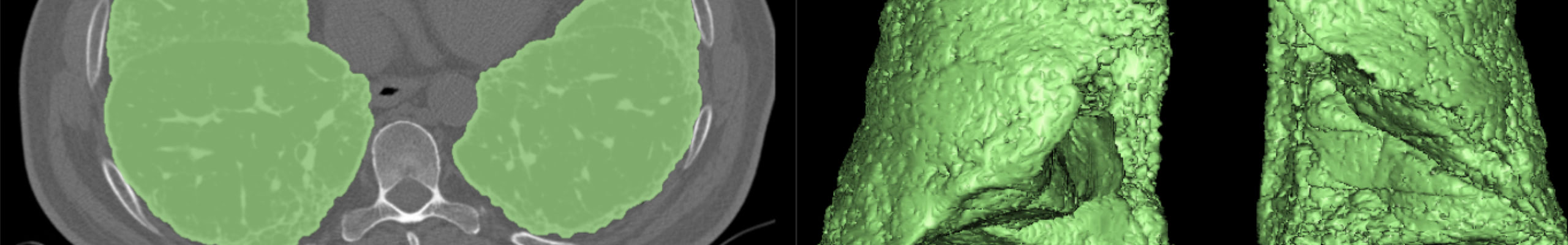
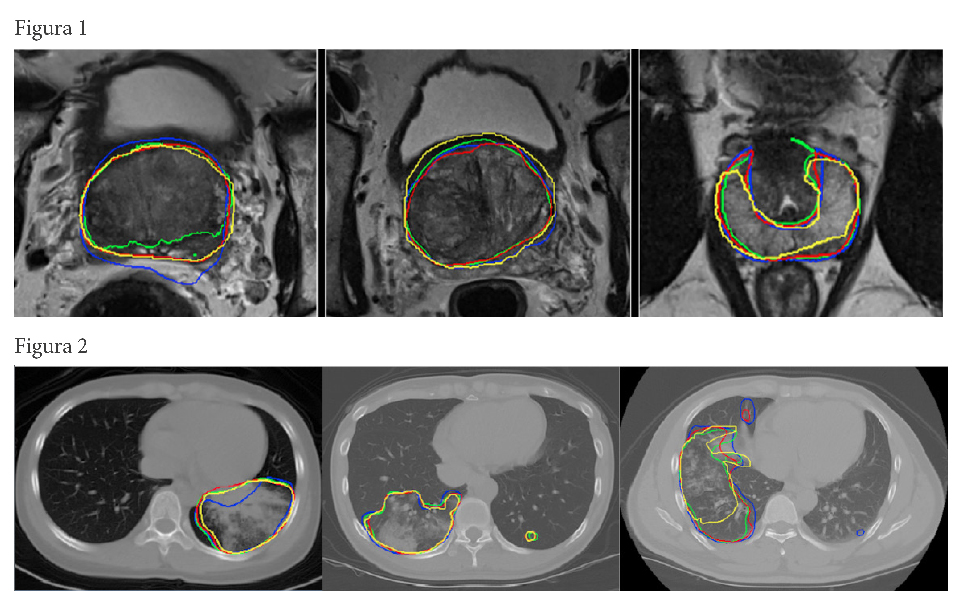
– A deep learning framework for segmentation of the whole-gland and zonal prostate, of the aneurysmal aorta and its valve but also parenchyma with idiopathic pulmonary fibrosis that provide accurate and fast segmentation results after being trained with a small image dataset of High-Resolution Computerized Tomography.
Pipeline
-
CLINICAL
NEED -
DISEASES
ANALYSIS - DISCOVERY
-
PRECLINICAL
VALIDATION -
PRECLINICAL
DEVELOPMENT -
CLINICAL
STUDIES
Principal Investigator
Contact
Therapeutic Areas:
Product:
Biomarkers, Medical Devices & Tissue Engineering
Collaborations:
- Istituto Mediterraneo per i Trapianti (ISMETT) IRCCS, Palermo, Italia
- Istituto di Bioimmagini e Fisiologia Molecolare (IBFM-CNR), Cefalù, Italia
- Georgia Institute of Technology (GIT), Atlanta, USA
- Unità di Medicina Nucleare, Università di Messina, Messina, Italia
- Unità di Medicina Nucleare, Fondazione Istituto G.Giglio, Cefalù, Italia
- Dipartimento di Ingegneria, Università di Palermo, Palermo, Italia
- Unità di Fisica Medica, Ospedale Cannizzaro, Catania, Italia
- Dipartimento di Medicina Nucleare, Ospedale Cannizzaro, Catania, Italia
- Dipartimento di Medicina e Chirurgia Clinica, Università degli Studi di Napoli “Federico II”, Napoli, Italia
Scarica il pdf del progetto